Difference between revisions of "Analysis Package"
(→Automated Segmentation) |
(→Recommended Analysis Packages) |
||
| (15 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| − | == Overview == | + | == Recommended Analysis Packages == |
| + | We currently recommend using the following combination of packages for Miniscope analysis | ||
| + | *Image Registration: [https://github.com/flatironinstitute/NoRMCorre NoRMCorre] or [https://scanbox.wordpress.com/2014/03/20/recursive-image-alignment-and-statistics/ Dario Ringach's recursive image registration algorithm]. NoRMCorre can correct for both local and global rigid motion while Dario's approach works just on a global level. | ||
| + | *Neuron segmentation and activity extraction: [https://github.com/zhoupc/CNMF_E CNMF-E] works extremely well for Miniscope imaging. It can denoise, demix, and deconvolve neural activity. | ||
| + | *Tracking neurons across days: [https://github.com/zivlab/CellReg CellReg] | ||
| + | |||
| + | Luckily, multiple groups are working on analysis packages which combine the approaches listed above into a single workflow: | ||
| + | *[https://github.com/flatironinstitute/CaImAn CaImAn]: "Computational toolbox for large scale Calcium Imaging Analysis, including movie handling, motion correction, source extraction, spike deconvolution and result visualization." This toolbox is very well maintained by the Simons Foundation and generally has the most up-to-date versions of NoRMCorre and CNMF. | ||
| + | *[https://github.com/PeyracheLab/miniscoPy MiniscoPy]: "A package to analyse calcium imaging data recorded with the Miniscope." This package is being developed by Adrien Peyrache's lab at McGill and is based on the CaImAn toolbox. | ||
| + | *[https://github.com/etterguillaume/MiniscopeAnalysis MiniscopeAnalysis]: "Analysis package for miniscope data Written by Guillaume Etter (Sylvain Williams Lab, McGill University) This is an updated version of the Miniscope analysis package developed by Daniel Aharoni at UCLA in collaboration with the Golshani lab, Silva lab, and Khaki lab. It combines core functions of the original ms analysis package with NormCorre alignment and CNMFE source extraction." | ||
| + | |||
| + | A new approach, as of June 2018, has come out for analyzing 1 photon imaging data. While we haven't tested this pipeline ourselves yet, it looks extremely promising. | ||
| + | *[https://github.com/JinghaoLu/MIN1PIPE MIN1PIPE]: "MIN1PIPE is a fully automatic, Matlab-based toolbox, solving the full range problems in 1-photon calcium imaging in one package: data enhancement → movement morrection → signal extraction. It requires minimal parameter-tuning and integrates the semi-auto options. Each inidividual module can also be easily adapted for the 2-photon imaging setting." | ||
| + | * Associated paper can be found [https://www.biorxiv.org/content/early/2018/04/30/311548 here]. | ||
| + | |||
| + | == Old Analysis Package Description Below (not recommended) == | ||
| + | This information below refers to the original Miniscope analysis package we developed. While we still provide this analysis package on GitHub we highly recommend not using it and instead using one of the analysis packages listed above. | ||
| + | === Overview === | ||
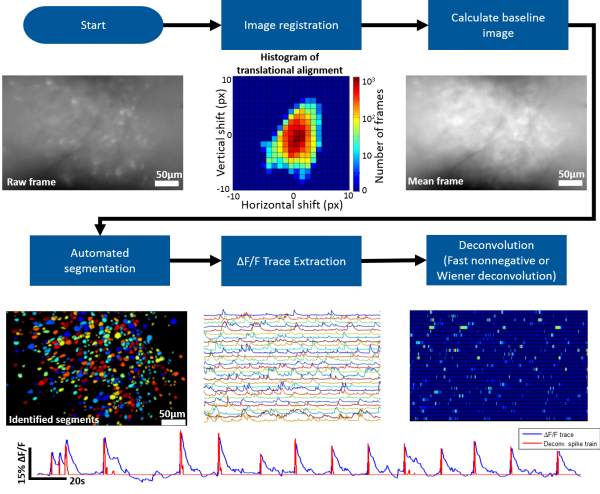
[[File:AnalysisOverview.png|thumb|600px]] | [[File:AnalysisOverview.png|thumb|600px]] | ||
The MATLAB analysis package provided on this wiki contains the tools necessary for going from the raw microscope video to deconvolved individual neuron activity. Our package supports batch (generally run in parallel) processing for handling multiple recording sessions and can also be run on an individual session stored in the MATLAB workspace. | The MATLAB analysis package provided on this wiki contains the tools necessary for going from the raw microscope video to deconvolved individual neuron activity. Our package supports batch (generally run in parallel) processing for handling multiple recording sessions and can also be run on an individual session stored in the MATLAB workspace. | ||
| Line 23: | Line 40: | ||
<br clear=all> | <br clear=all> | ||
| − | == Work Flow == | + | === Work Flow === |
| − | A general work flow for processing and analyzing miniscope data can be found in msBatchRun.m. This script will walk you through all the necessary functions to go from the raw data to approximate spiking activity of individual neurons. | + | A general work flow for processing and analyzing miniscope data can be found in msBatchRun.m for batch processing and msRun.m for single recording processing. This script will walk you through all the necessary functions to go from the raw data to approximate spiking activity of individual neurons. We suggest starting with msRun.m and walking through all steps for a single recording session. You can use '''msPlayVidObj'''(vidObj,downSamp,columnCorrect, align, dFF, overlay) to display a video of your data while applying certain aspect of the data processing workflow. An example use of this function to display an aligned dF/F video downsampled by a factor of 5 would be '''msPlayVidObj'''(ms,5,true,true,true,false). |
| + | |||
| + | {{#ev:youtube|https://youtu.be/7paBLG7aFIo|640|center}} | ||
| − | == Image Registration == | + | === Image Registration === |
We suggest using [https://scanbox.wordpress.com/2014/03/20/recursive-image-alignment-and-statistics/ Dario Ringach's recursive image registration algorithm]. | We suggest using [https://scanbox.wordpress.com/2014/03/20/recursive-image-alignment-and-statistics/ Dario Ringach's recursive image registration algorithm]. | ||
<br clear=all> | <br clear=all> | ||
| − | == Automated Segmentation == | + | === Automated Segmentation === |
| + | Our MATLAB analysis package implements an novel fully automated segmentation algorithm. Roughly speaking the algorithm follows the following steps: | ||
| + | *Detect ΔF/F local maxima throughout recording and record their spatiotemporal location. | ||
| + | *For each spatial location where local maxima occurred, generate a pixel group, G_px, containing pixels at the center of this location. | ||
| + | *Calculate the cross correlation of G_px’s mean ΔF/F trace with the individual ΔF/F traces of surrounding pixels, limiting the ΔF/F traces to only the frames temporally close to nearby local maxima events. | ||
| + | *Pixels with a high correlation coefficient are added to G_px. | ||
| + | *Repeat until an iteration occurs where no more surrounding pixels are added to G_px. | ||
| + | <br clear=all> | ||
| − | + | === dF/F Trace Extraction and Deconvolution === | |
| − | == dF/F Trace Extraction and Deconvolution == | + | After segments (ROIs of each active neuron) have been identified dF/F traces can be extracted then deconvolved to approximate spiking activity of each segment. We prefer the the non-negative deconvolution approach of [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3007657/ Vogelstein et. al.]. |
| − | == Alignment Across Sessions == | + | === Alignment Across Sessions === |
Latest revision as of 12:10, 21 June 2018
Contents
Recommended Analysis Packages
We currently recommend using the following combination of packages for Miniscope analysis
- Image Registration: NoRMCorre or Dario Ringach's recursive image registration algorithm. NoRMCorre can correct for both local and global rigid motion while Dario's approach works just on a global level.
- Neuron segmentation and activity extraction: CNMF-E works extremely well for Miniscope imaging. It can denoise, demix, and deconvolve neural activity.
- Tracking neurons across days: CellReg
Luckily, multiple groups are working on analysis packages which combine the approaches listed above into a single workflow:
- CaImAn: "Computational toolbox for large scale Calcium Imaging Analysis, including movie handling, motion correction, source extraction, spike deconvolution and result visualization." This toolbox is very well maintained by the Simons Foundation and generally has the most up-to-date versions of NoRMCorre and CNMF.
- MiniscoPy: "A package to analyse calcium imaging data recorded with the Miniscope." This package is being developed by Adrien Peyrache's lab at McGill and is based on the CaImAn toolbox.
- MiniscopeAnalysis: "Analysis package for miniscope data Written by Guillaume Etter (Sylvain Williams Lab, McGill University) This is an updated version of the Miniscope analysis package developed by Daniel Aharoni at UCLA in collaboration with the Golshani lab, Silva lab, and Khaki lab. It combines core functions of the original ms analysis package with NormCorre alignment and CNMFE source extraction."
A new approach, as of June 2018, has come out for analyzing 1 photon imaging data. While we haven't tested this pipeline ourselves yet, it looks extremely promising.
- MIN1PIPE: "MIN1PIPE is a fully automatic, Matlab-based toolbox, solving the full range problems in 1-photon calcium imaging in one package: data enhancement → movement morrection → signal extraction. It requires minimal parameter-tuning and integrates the semi-auto options. Each inidividual module can also be easily adapted for the 2-photon imaging setting."
- Associated paper can be found here.
Old Analysis Package Description Below (not recommended)
This information below refers to the original Miniscope analysis package we developed. While we still provide this analysis package on GitHub we highly recommend not using it and instead using one of the analysis packages listed above.
Overview
The MATLAB analysis package provided on this wiki contains the tools necessary for going from the raw microscope video to deconvolved individual neuron activity. Our package supports batch (generally run in parallel) processing for handling multiple recording sessions and can also be run on an individual session stored in the MATLAB workspace.
Roughly speaking the steps involved are the following
- Generate MATLAB data structure
- All access to the raw or processed data is handled through this data structure
- Memory efficient (less than 10MB per minute of recording)
- Correct for motion artifacts using an amplitude or FFT image registration algorithm
- Identify the bountries of individual neurons (we provide a novel, fully automated segmentation approach)
- Extract dF/F activity for identified neurons
- Deconvolve dF/F activity using Wiener or Fast Nonnegative deconvolution to approximate neuronal spiking pattern
- Additional Analysis:
- Cross-talk removal
- Segment matching across sessions
- Behavior tracking and syncing to microscope data
- Activity detection
The Miniscope Analysis Github repository can be found here.
Work Flow
A general work flow for processing and analyzing miniscope data can be found in msBatchRun.m for batch processing and msRun.m for single recording processing. This script will walk you through all the necessary functions to go from the raw data to approximate spiking activity of individual neurons. We suggest starting with msRun.m and walking through all steps for a single recording session. You can use msPlayVidObj(vidObj,downSamp,columnCorrect, align, dFF, overlay) to display a video of your data while applying certain aspect of the data processing workflow. An example use of this function to display an aligned dF/F video downsampled by a factor of 5 would be msPlayVidObj(ms,5,true,true,true,false).
Image Registration
We suggest using Dario Ringach's recursive image registration algorithm.
Automated Segmentation
Our MATLAB analysis package implements an novel fully automated segmentation algorithm. Roughly speaking the algorithm follows the following steps:
- Detect ΔF/F local maxima throughout recording and record their spatiotemporal location.
- For each spatial location where local maxima occurred, generate a pixel group, G_px, containing pixels at the center of this location.
- Calculate the cross correlation of G_px’s mean ΔF/F trace with the individual ΔF/F traces of surrounding pixels, limiting the ΔF/F traces to only the frames temporally close to nearby local maxima events.
- Pixels with a high correlation coefficient are added to G_px.
- Repeat until an iteration occurs where no more surrounding pixels are added to G_px.
dF/F Trace Extraction and Deconvolution
After segments (ROIs of each active neuron) have been identified dF/F traces can be extracted then deconvolved to approximate spiking activity of each segment. We prefer the the non-negative deconvolution approach of Vogelstein et. al..